import geopandas as gpdimport pandas as pd# random coordinates = gpd.GeoDataFrame(geometry= gpd.points_from_xy([0 , 0 , 0 ], [0 , 90 , 120 ]))= gpd.GeoDataFrame(geometry= gpd.points_from_xy([0 , 0 ], [0 , - 90 ]))apply (lambda g: gdf_2.distance(g))
0
0.0
90.0
1
90.0
180.0
2
120.0
210.0
apply (lambda g: gdf_1.distance(g))
0
0.0
90.0
120.0
1
90.0
0.0
30.0
2
120.0
30.0
0.0
apply (lambda g: gdf_1.distance(g)).median(axis= 0 )
0 90.0
1 30.0
2 30.0
dtype: float64
= gpd.read_file('/home/jupyter-student/behavioralHealth.shp' )
ERROR 1: PROJ: proj_create_from_database: Open of /opt/tljh/user/share/proj failed
= gpd.read_parquet('/home/jupyter-student/data/scag_region.parquet' )
<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World.
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984 ensemble
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
= socal.to_crs(clinics.crs)
= socal[socal.geoid.str .startswith("06065" )]
apply (lambda g: clinics.distance(g)).median(axis= 0 )
0 104070.136497
1 110145.422279
2 118486.478639
3 780106.703273
4 109999.756757
5 110145.422279
6 93068.512487
7 127360.564230
8 110274.481746
9 130577.585244
10 110145.422279
11 303743.579448
12 110145.422279
13 162687.749002
14 110501.825357
15 226157.792611
16 130575.090996
17 86833.101138
18 128491.121930
19 162684.638468
20 145278.329356
21 226161.334468
22 299007.725394
23 105090.401115
24 110146.607263
25 104067.484478
26 110501.616968
27 110499.112055
dtype: float64
apply (lambda g: rc.distance(g)).median(axis= 0 )
171 35414.497106
172 57404.024199
173 70444.292093
174 59916.246444
175 65899.907435
...
4559 51754.814636
4560 69235.098968
4568 121785.198080
4569 147379.220664
4578 261177.893498
Length: 453, dtype: float64
apply (lambda g: rc.distance(g)).min (axis= 0 )
171 17587.507322
172 0.000000
173 11703.070730
174 6093.825933
175 10885.748131
...
4559 23468.056401
4560 14504.796947
4568 45895.492409
4569 3988.154669
4578 10834.765951
Length: 453, dtype: float64
'cent' ] = clinics.centroid
/opt/tljh/user/lib/python3.10/site-packages/geopandas/geodataframe.py:1543: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
super().__setitem__(key, value)
'cent' , inplace= True )
apply (lambda g: rc.distance(g)).min (axis= 0 )
171 23599.208237
172 3019.158172
173 14930.591185
174 7500.806546
175 17330.260026
...
4559 27631.243530
4560 16783.956907
4568 50460.359167
4569 6795.067162
4578 14591.568700
Length: 453, dtype: float64
'dist' ] = clinics.geometry.apply (lambda g: rc.distance(g)).min (axis= 0 )
/opt/tljh/user/lib/python3.10/site-packages/geopandas/geodataframe.py:1543: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
super().__setitem__(key, value)
'geometry' , inplace= True )
171 41397.573034
172 19270.082397
173 24544.962547
174 12246.346442
175 16792.718165
...
4559 19985.523408
4560 34261.675094
4568 25011.033708
4569 14872.897004
4578 22086.110487
Name: per_capita_income, Length: 453, dtype: float64
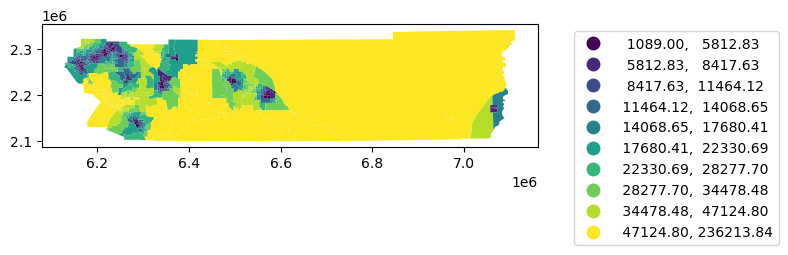
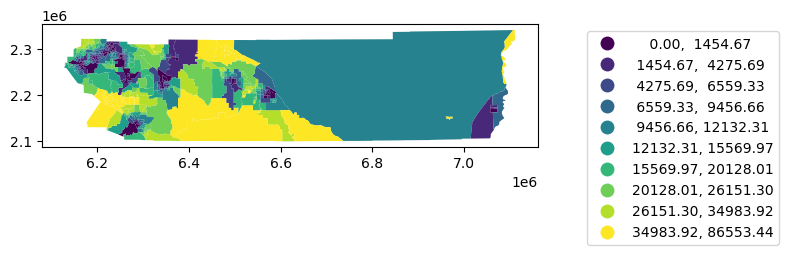
= 'dist' , scheme= 'quantiles' ,k= 10 , legend= True ,= {'bbox_to_anchor' : (1.5 , 1 )})
Alternative with sjoin
First with polygon to clinic
= gpd.sjoin_nearest(rc, clinics, distance_col= 'distance_to_clinic' )
'geoid' , 'distance_to_clinic' ]].groupby(by= 'geoid' ).min ()
geoid
06065030101
1768.344059
06065030103
3928.348106
06065030104
3284.836973
06065030200
7529.946188
06065030300
5266.871283
...
...
06065941300
29557.523443
06065941400
22685.975067
06065941500
14832.118828
06065980004
9830.995241
06065981000
86553.444747
453 rows × 1 columns
= nq[['geoid' , 'distance_to_clinic' ]].groupby(by= 'geoid' ).max ()= True )
0
06065030101
1768.344059
1
06065030103
3928.348106
2
06065030104
3284.836973
3
06065030200
7529.946188
4
06065030300
5266.871283
= rc.merge(nq, left_on= 'geoid' , right_on= 'geoid' )'distance_to_clinic' , scheme= 'quantiles' , k= 10 , legend= True ,= {'bbox_to_anchor' : (1.5 , 1 )});
now on centroids of tracts
171 POINT (6228927.025 2271856.585)
172 POINT (6232768.433 2305432.441)
173 POINT (6267251.000 2290261.360)
174 POINT (6257557.755 2288526.028)
175 POINT (6263055.600 2265924.366)
...
4559 POINT (6174619.367 2298965.242)
4560 POINT (6159739.434 2285871.688)
4568 POINT (6286719.240 2191439.849)
4569 POINT (6348755.310 2217649.506)
4578 POINT (6485172.924 2237980.191)
Length: 453, dtype: geometry
'centroid' ] = rc.centroid= rc.set_geometry('centroid' )
/opt/tljh/user/lib/python3.10/site-packages/geopandas/geodataframe.py:1543: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
super().__setitem__(key, value)
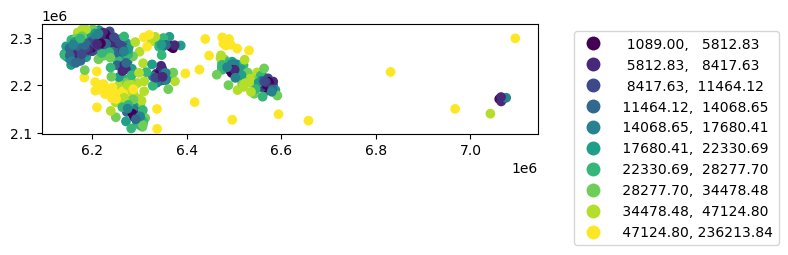
= gpd.sjoin_nearest(rc, clinics, distance_col= 'distance_to_clinic' )
'geoid' , 'distance_to_clinic' ]].groupby(by= 'geoid' ).min ()
geoid
06065030101
2532.133821
06065030103
6435.915812
06065030104
7886.872071
06065030200
13082.177204
06065030300
10765.815358
...
...
06065941300
33303.132676
06065941400
26309.841401
06065941500
29645.998724
06065980004
17580.079738
06065981000
94440.398999
453 rows × 1 columns
= nq[['geoid' , 'distance_to_clinic' ]].groupby(by= 'geoid' ).min ()
= True )
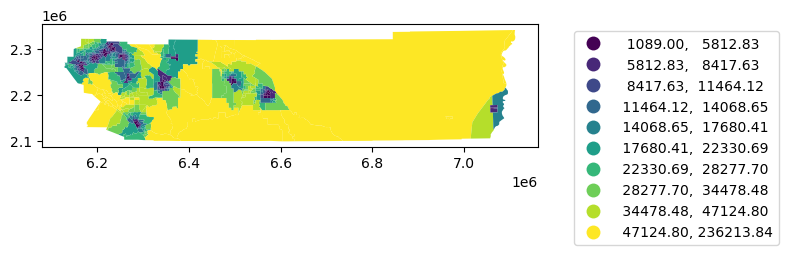
= rc.merge(nq, left_on= 'geoid' , right_on= 'geoid' )'distance_to_clinic' , scheme= 'quantiles' , k= 10 , legend= True ,= {'bbox_to_anchor' : (1.5 , 1 )})
= res.set_geometry('geometry' )
'distance_to_clinic' , scheme= 'quantiles' , k= 10 , legend= True ,= {'bbox_to_anchor' : (1.5 , 1 )})
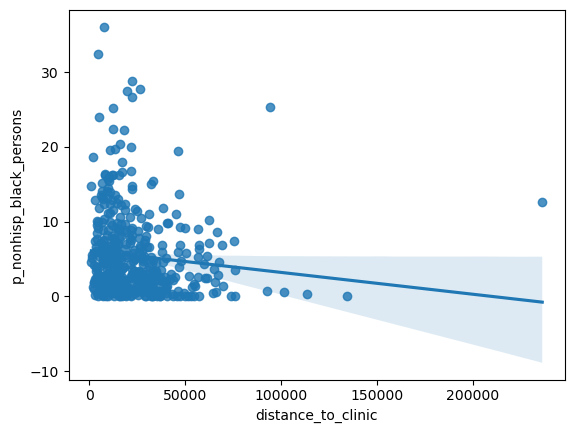
= seaborn.regplot(x= 'distance_to_clinic' , y= 'p_nonhisp_black_persons' , data= res)
'distance_to_clinic' , 'per_capita_income' , 'p_nonhisp_black_persons' ]].corr()
distance_to_clinic
1.000000
0.119891
-0.108220
per_capita_income
0.119891
1.000000
-0.235644
p_nonhisp_black_persons
-0.108220
-0.235644
1.000000
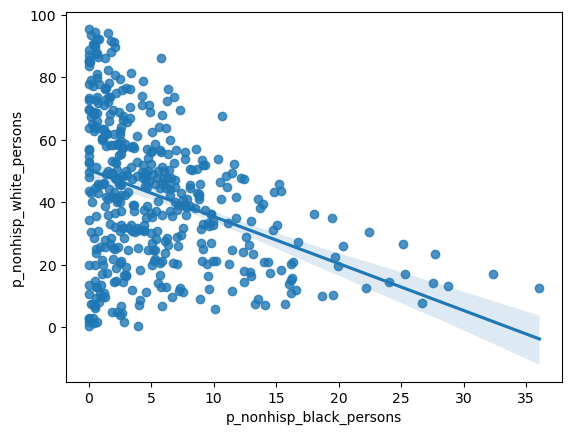
= seaborn.regplot(x= 'distance_to_clinic' , y= 'p_nonhisp_white_persons' , data= res)
= seaborn.regplot(x= 'p_nonhisp_black_persons' , y= 'p_nonhisp_white_persons' , data= res)
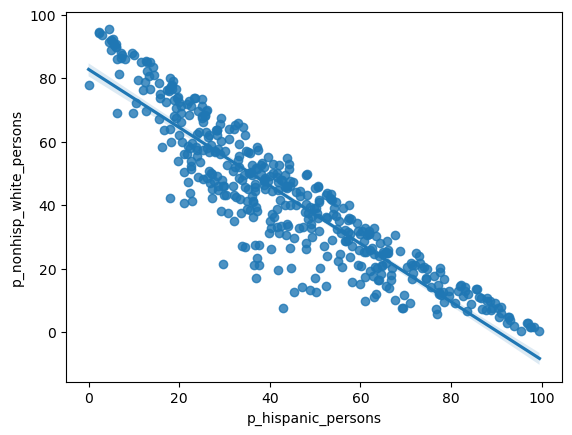
= seaborn.regplot(x= 'p_hispanic_persons' , y= 'p_nonhisp_white_persons' , data= rc)
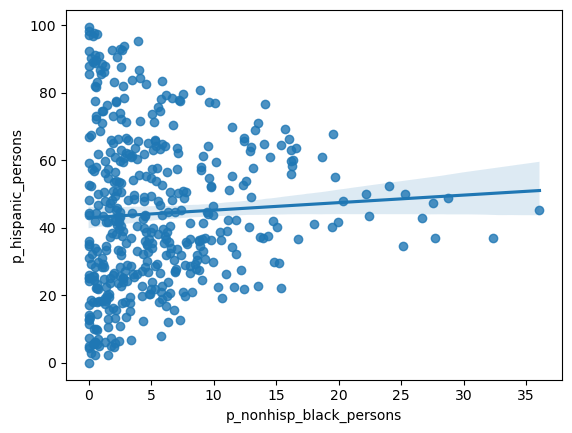
= seaborn.regplot(x= 'p_nonhisp_black_persons' , y= 'p_hispanic_persons' , data= rc)