from geosnap import DataStore
import geopandas as gpd
datasets = DataStore()
from geosnap.io import get_acs
ca = get_acs(datasets, state_fips=['06'], level='tract', years=[2016])
sd = ca[ca.geoid.str.startswith('06073')]
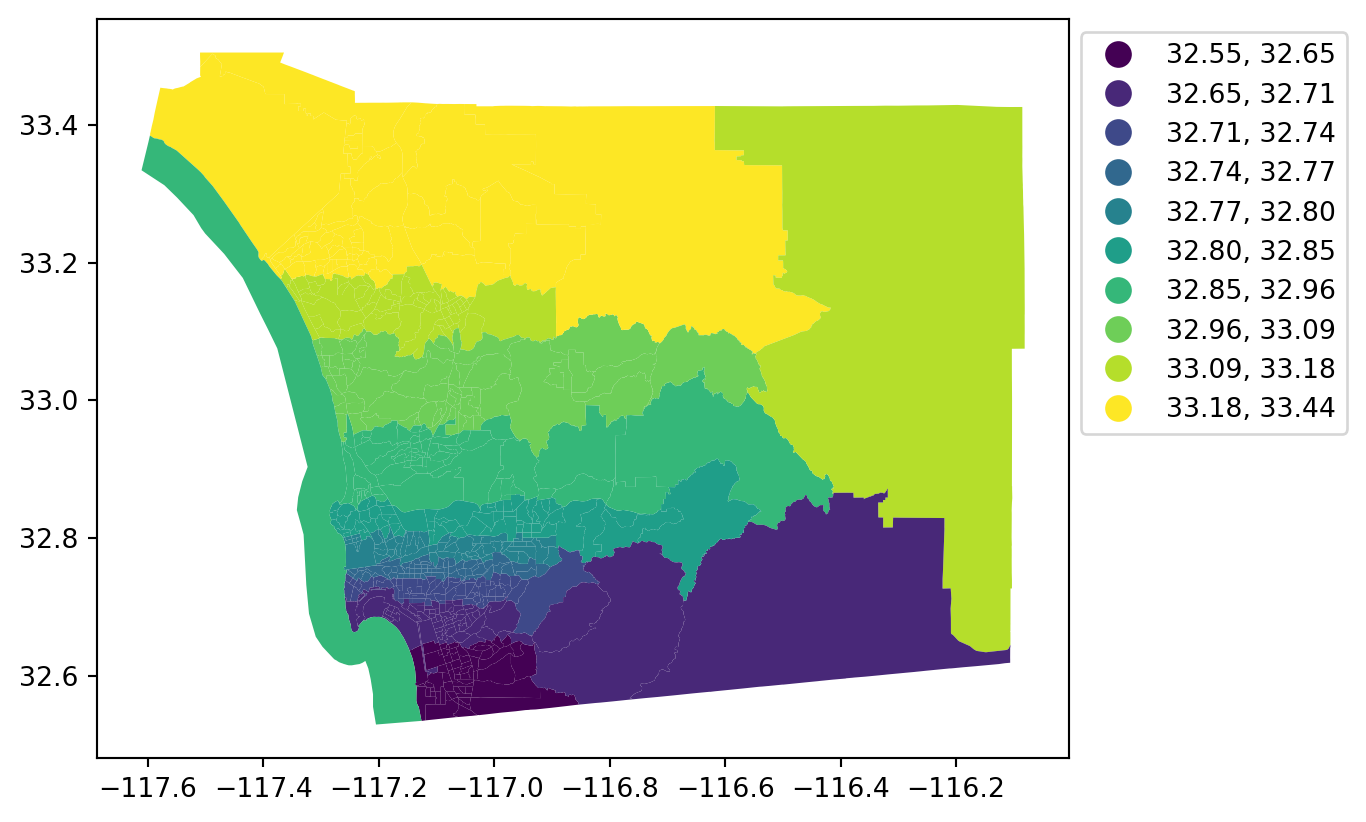
sd['lat'] = sd.geometry.centroid.y
sd.plot('lat', scheme='quantiles', k=10, legend=True,
legend_kwds={'bbox_to_anchor': (1.3,1)});